النبات

مواضيع عامة في علم النبات

الجذور - السيقان - الأوراق

النباتات الوعائية واللاوعائية

البذور (مغطاة البذور - عاريات البذور)

الطحالب

النباتات الطبية


الحيوان

مواضيع عامة في علم الحيوان

علم التشريح

التنوع الإحيائي

البايلوجيا الخلوية


الأحياء المجهرية

البكتيريا

الفطريات

الطفيليات

الفايروسات


علم الأمراض

الاورام

الامراض الوراثية

الامراض المناعية

الامراض المدارية

اضطرابات الدورة الدموية

مواضيع عامة في علم الامراض

الحشرات


التقانة الإحيائية

مواضيع عامة في التقانة الإحيائية


التقنية الحيوية المكروبية

التقنية الحيوية والميكروبات

الفعاليات الحيوية

وراثة الاحياء المجهرية

تصنيف الاحياء المجهرية

الاحياء المجهرية في الطبيعة

أيض الاجهاد

التقنية الحيوية والبيئة

التقنية الحيوية والطب

التقنية الحيوية والزراعة

التقنية الحيوية والصناعة

التقنية الحيوية والطاقة

البحار والطحالب الصغيرة

عزل البروتين

هندسة الجينات


التقنية الحياتية النانوية

مفاهيم التقنية الحيوية النانوية

التراكيب النانوية والمجاهر المستخدمة في رؤيتها

تصنيع وتخليق المواد النانوية

تطبيقات التقنية النانوية والحيوية النانوية

الرقائق والمتحسسات الحيوية

المصفوفات المجهرية وحاسوب الدنا

اللقاحات

البيئة والتلوث


علم الأجنة

اعضاء التكاثر وتشكل الاعراس

الاخصاب

التشطر

العصيبة وتشكل الجسيدات

تشكل اللواحق الجنينية

تكون المعيدة وظهور الطبقات الجنينية

مقدمة لعلم الاجنة


الأحياء الجزيئي

مواضيع عامة في الاحياء الجزيئي


علم وظائف الأعضاء


الغدد

مواضيع عامة في الغدد

الغدد الصم و هرموناتها

الجسم تحت السريري

الغدة النخامية

الغدة الكظرية

الغدة التناسلية

الغدة الدرقية والجار الدرقية

الغدة البنكرياسية

الغدة الصنوبرية

مواضيع عامة في علم وظائف الاعضاء

الخلية الحيوانية

الجهاز العصبي

أعضاء الحس

الجهاز العضلي

السوائل الجسمية

الجهاز الدوري والليمف

الجهاز التنفسي

الجهاز الهضمي

الجهاز البولي


المضادات الميكروبية

مواضيع عامة في المضادات الميكروبية

مضادات البكتيريا

مضادات الفطريات

مضادات الطفيليات

مضادات الفايروسات

علم الخلية

الوراثة

الأحياء العامة

المناعة

التحليلات المرضية

الكيمياء الحيوية

مواضيع متنوعة أخرى

الانزيمات
Transfer of T-DNA Resembles Bacterial Conjugation
المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
المصدر:
LEWIN’S GENES XII
الجزء والصفحة:
10-4-2021
2307
Transfer of T-DNA Resembles Bacterial Conjugation
KEY CONCEPTS
- T-DNA is generated when a nick at the right boundary creates a primer for synthesis of a new DNA strand.
- The preexisting single strand that is displaced by the new synthesis is transferred to the plant cell nucleus.
- Transfer is terminated when DNA synthesis reaches a nick at the left boundary.
- The T-DNA is transferred as a complex of singlestranded DNA with the VirE2 single-strand binding protein.
- The single-stranded T-DNA is converted into doublestranded DNA and integrated into the plant genome.
- The mechanism of integration is not known. T-DNA can be used to transfer genes into a plant nucleus.
The transfer process actually selects the T-region for entry into the plant. FIGURE 1 shows that the T-DNA of a nopaline plasmid is demarcated from the flanking regions in the Ti plasmid by repeats of 25 bp, which differ at only two positions between the left and right ends. When T-DNA is integrated into a plant genome, it has a well-defined right junction, which retains 1 to 2 bp of the right repeat. The left junction is variable; the boundary of T-DNA in the plant genome can be located at the 25-bp repeat or at one of a series of sites extending over about 100 bp within the T-DNA. At times multiple tandem copies of T-DNA are integrated at a single site.
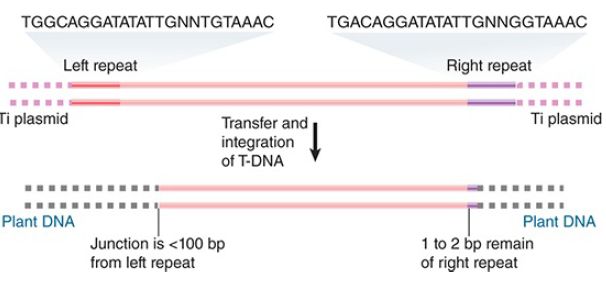
FIGURE 1.T-DNA has almost identical repeats of 25 bp at each end in the Ti plasmid. The right repeat is necessary for transfer and integration to a plant genome. T-DNA that is integrated in a plant genome has a precise junction that retains 1 to 2 bp of the right repeat, but the left junction varies and may be up to 100 bp short of the left repeat.
The virD locus has four ORFs. Two of the proteins encoded at virD —VirD1 and VirD2—provide an endonuclease that initiates the transfer process by nicking T-DNA at a specific site. FIGURE 2. illustrates a model for transfer. A nick is made at the right 25-bp repeat. It provides a priming end for synthesis of a DNA single strand. Synthesis of the new strand displaces the old strand, which is used in the transfer process. Transfer is terminated when DNA synthesis reaches a nick at the left repeat. This model explains why the right repeat is essential, and it accounts for the polarity of the process. If the left repeat fails to be nicked, transfer could continue farther along the Ti plasmid.
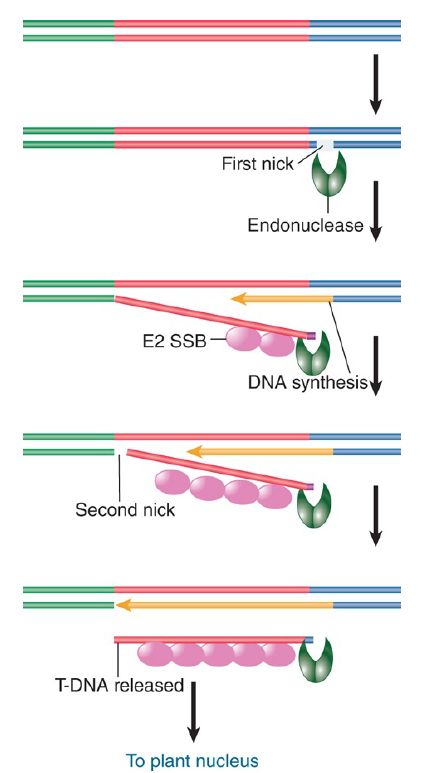
FIGURE 2. T-DNA is generated by displacement when DNA synthesis starts at a nick made at the right repeat. The reaction is terminated by a nick at the left repeat.
The transfer process involves production of a single molecule of single-stranded DNA in the infecting bacterium. It is transferred in the form of a DNA–protein complex, sometimes called the Tcomplex.
The DNA is covered by the VirE2 single-strand binding protein, which has a nuclear localization signal and is responsible for transporting T-DNA into the plant cell nucleus. A single molecule of the D2 subunit of the endonuclease remains bound at the 5′ end.
The virB operon codes for 11 products that are involved in the transfer reaction.
Outside T-DNA, immediately adjacent to the right border, is another short sequence called overdrive, which greatly stimulates the transfer process. Overdrive functions like an enhancer: It must lie on the same molecule of DNA, but enhances the efficiency of transfer even when located several thousand base pairs away from the border. VirC1, and possibly VirC2, may act at the overdrive sequence.
Octopine plasmids have a more complex pattern of integrated TDNA than nopaline plasmids. The pattern of T-strands is also more complex, and several discrete species can be found, corresponding to elements of T-DNA. This suggests that octopine T-DNA has several sequences that provide targets for nicking and/or termination of DNA synthesis.
This model for transfer of T-DNA closely resembles the events involved in bacterial conjugation, when the E. coli chromosome is transferred from one cell to another in single-stranded form. The genes of the virB operon are homologous to the tra genes of certain bacterial plasmids (including the tra operons on Ti-plasmids) that are involved in conjugation . Together with VirD4 (a coupling protein), the gene products of the virB genes form a T4SS.
The T strand, along with several other Vir proteins, is then exported into the plant cell by the T4SS, a step that requires interaction of the bacterial T-pilus with at least one host-specific protein. The Tstrand molecule is coated with numerous VirE2 molecules when entering the plant-cell cytoplasm. These molecules confer to the TDNA the structure and protection needed for its travel to the plantcell nucleus .
Researchers do not know how the transferred DNA is integrated into the plant genome. At some stage, the newly generated single strand must be converted into duplex DNA. Circles of T-DNA that are found in infected plant cells appear to be generated by recombination between the left and right 25-bp repeats, but researchers do not know if they are intermediates. The actual event is likely to involve nonhomologous recombination, because there is no homology between the T-DNA and the sites of integration.
What is the structure of the target site? Sequences flanking the integrated T-DNA tend to be rich in A-T base pairs (a feature displayed in target sites for some transposable elements). The sequence rearrangements that occur at the ends of the integrated T-DNA make it difficult to analyze the structure. Researchers do not know whether the integration process generates new sequences in the target DNA comparable to the target repeats created in transposition.
T-DNA is expressed at its site of integration. The region contains several transcription units, each of which probably contains a gene expressed from an individual promoter. Their functions are concerned with the state of the plant cell, maintaining its tumorigenic properties, controlling shoot and root formation, and suppressing differentiation into other tissues. None of these genes is needed for T-DNA transfer.
The Ti plasmid presents an interesting organization of functions. Outside the T-region, it carries genes needed to initiate oncogenesis; at least some are concerned with the transfer of TDNA, and researchers would like to know whether others function in the plant cell to affect its behavior at this stage. Also outside the T-region are the genes that enable the Agrobacterium to catabolize the opine that the transformed plant cell will produce. Within the Tregion are the genes that control the transformed state of the plant as well as the genes that cause it to synthesize the opines that will benefit the Agrobacterium that originally provided the T-DNA.
As a practical matter, the ability of Agrobacterium to transfer TDNA to the plant genome makes it possible to introduce new genes into plants. The transfer/integration and oncogenic functions are separate; thus, it is possible to engineer new Ti plasmids in which the oncogenic functions have been replaced by other genes whose effect on the plant researchers wish to test. The existence of a natural system for delivering genes to the plant genome has greatly facilitated genetic engineering of plants.
 الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة

الآخبار الصحية















 قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة
قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة "المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة
"المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة (نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)
(نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)