
Ribosomal RNA Is Found Throughout Both Ribosomal Subunits
 المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
 المصدر:
LEWIN’S GENES XII
المصدر:
LEWIN’S GENES XII
 الجزء والصفحة:
الجزء والصفحة:
 25-5-2021
25-5-2021
 2161
2161
Ribosomal RNA Is Found Throughout Both Ribosomal Subunits
KEY CONCEPTS
- Each rRNA has several distinct domains that fold independently.
- Virtually all ribosomal proteins are in contact with rRNA.
- Most of the contacts between ribosomal subunits are made between the 16S and 23S rRNAs.
Two-thirds of the mass of the bacterial ribosome is made up of rRNA. The most revealing approach to analyzing secondary structure of large RNAs is to compare the sequences of homologous rRNAs in related organisms. Those regions that are important in the secondary structure retain the ability to interact by base pairing. Thus, if a base pair is required, it can form at the same relative position in each rRNA. This approach has enabled detailed models of 16S and 23S rRNA to be constructed.
Each of the major rRNAs has a secondary structure with several discrete domains. Four general domains are formed by 16S rRNA, in which just under half of the sequence is base paired. Six general domains are formed by 23S rRNA. The individual double-helical regions tend to be short (fewer than 8 bp). Frequently the duplex regions are not perfect and contain bulges of unpaired bases.
Comparable models have been drawn for mitochondrial rRNAs (which are shorter and have fewer domains) and for eukaryotic cytosolic rRNAs (which are longer and have more domains). The greater length of eukaryotic rRNAs is due largely to the acquisition of sequences representing additional domains. The crystal structure of the ribosome shows that in each subunit the domains of the major rRNA fold independently and have discrete locations.
Differences in the ability of 16S rRNA to react with chemical agents are found when 30S subunits are compared with 70S ribosomes; there also are differences between separate ribosomal subunits and those engaged in translation. Changes in the reactivity of the rRNA occur when mRNA is bound, when the subunits associate, or when tRNA is bound. Some changes reflect a direct interaction of the rRNA with mRNA or tRNA, whereas others are caused indirectly by other changes in ribosome structure. The main point is that ribosome conformation is flexible during translation, particularly that of the small subunit, because it must physically check the accuracy of codon–anticodon pairing.
A feature of the primary structure of rRNA is the presence of methylated residues. There are about 10 methyl groups in 16S rRNA (located mostly toward the 3′ end of the molecule) and about 20 in 23S rRNA. In mammalian cells, the 18S and 28S rRNAs carry 43 and 74 methyl groups, respectively, so about 2% of the nucleotides are methylated (about three times the proportion of methylated nucleotides in bacterial rRNAs).
The large ribosomal subunit also contains a molecule of a 120-base 5S RNA (in all ribosomes except those of mitochondria). The sequence of 5S RNA is less well conserved than those of the major rRNAs. All 5S RNA molecules display a highly base-paired structure.
In eukaryotic cytosolic ribosomes, another small RNA is present in the large subunit, the 5.8S RNA. Its sequence corresponds to the 5′ end of the prokaryotic 23S rRNA.
Some ribosomal proteins bind strongly to isolated rRNAs. Others do not bind to free rRNAs, but can bind after other proteins have bound. This suggests that the conformation of the rRNA is important in determining whether binding sites exist for some proteins. As each protein binds, it induces conformational changes in the rRNA that make it possible for other proteins to bind. In E. coli, virtually all the 30S ribosomal proteins interact (albeit to varying degrees) with 16S rRNA. The binding sites on the proteins show a wide variety of structural features, suggesting that protein– RNA recognition mechanisms may be diverse.
The 70S ribosome has an asymmetric structure. Figure 1 shows a schematic of the structure of the 30S subunit, which is divided into four regions: the head, neck, body, and platform. Figure 2 shows a similar representation of the 50S subunit, where two prominent features are the central protuberance (where 5S rRNA is located) and the stalk (made of multiple copies of protein L7). Figure 3 shows that the platform of the small subunit fits into the notch of the large subunit. A cavity (resemblinga doughn ut, but not visible in the figure) between the subunits contains some of the important sites.
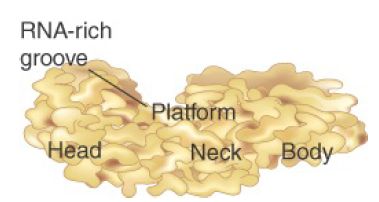
FIGURE 1 The 30S subunit has a head separated by a neck from the body, with a protruding platform.
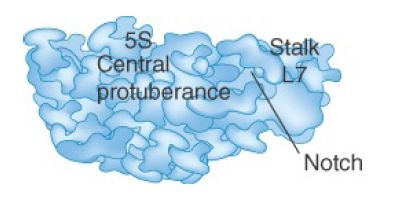
FIGURE 2 The 50S subunit has a central protuberance where 5S rRNA is located, separated by a notch from a stalk made of copies of the protein L7.
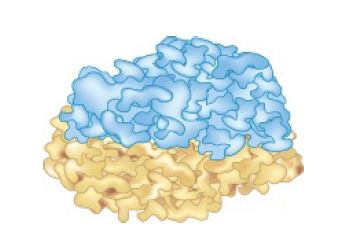
FIGURE 3 The platform of the 30S subunit fits into the notch of the 50S subunit to form the 70S ribosome.
The structure of the 30S subunit follows the organization of 16S rRNA, with each structural feature corresponding to a domain of the rRNA. The body is based on the 5′ domain, the platform on the central domain, and the head on the 3′ region. Figure 4 shows that the 30S subunit has an asymmetric distribution of RNA and protein. One important feature is that the platform of the 30S subunit that provides the interface with the 50S subunit is composed almost entirely of RNA. At most, two proteins (a small part of S7 and possibly part of S12) lie near the interface. This means that the association and dissociation of ribosomal subunits must depend on interactions with the 16S rRNA. Subunit association is affected by a mutation in a loop of 16S rRNA (at position 791) that is located at the subunit interface, and other nucleotides in 16S rRNA have been shown to be involved by modification/interference experiments. This observation supportsthe idea that the evolutionary origin of the ribosome may have been as a particle consisting solely of RNA rather than of both RNA protein.
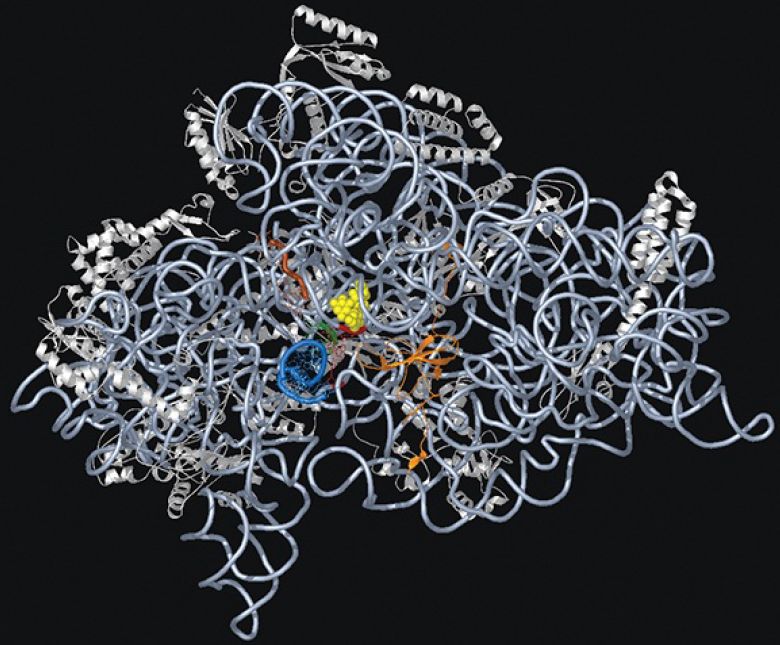
FIGURE 4. The 30S ribosomal subunit is a ribonucleoprotein particle. Ribosomal proteins are white and rRNA is light blue.
Courtesy of Dr. Kalju Kahn.
The 50S subunit has a more even distribution of components than the 30S does, with long rods of double-stranded RNA crisscrossing the structure. The RNA forms a mass of tightly packed helices. The exterior surface largely consists of protein, except for the peptidyl transferase center (see the section later in this chapter titled 23S rRNA Has Peptidyl Transferase Activity). Almost all segments of the 23S rRNA interact with protein, but many of the proteins are relatively unstructured.
The junction of subunits in the 70S ribosome involves contacts between 16S rRNA (many in the platform region) and 23S rRNA. A few interactions also occur between rRNAs of each subunit with proteins in the other and a few protein–protein contacts. Figure 22.40 identifies the contact points on the rRNA structures. Figure 22.41 opens out the structure (imagine the 50S subunit rotated counterclockwise and the 30S subunit rotated clockwise around the axis shown in the figure) to show the locations of the contact points on the face of each subunit.
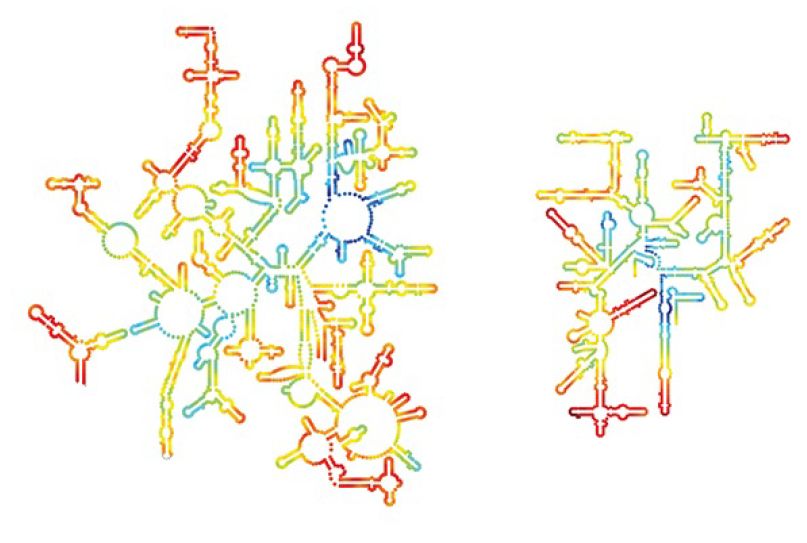
FIGURE 5. Contact points between the rRNAs are located in two domains of 16S rRNA and one domain of 23S rRNA.Laguna Design/Gett y Images.
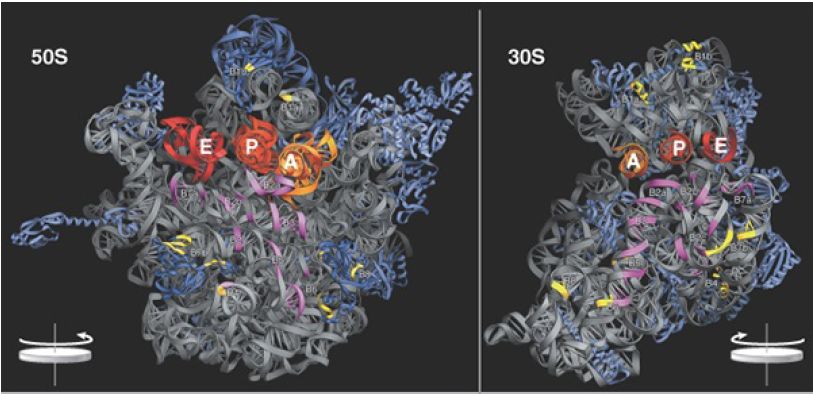
FIGURE 6. Contacts between the ribosomal subunits are mostly made by RNA (shown in purple). Contacts involving proteins are shown in yellow. The two subunits are rotated away from one another to show the faces where contacts are made; from a plane of contact perpendicular to the screen, the 50S subunit is rotated 90° counterclockwise, and the 30S is rotated 90° clockwise (this shows it in the reverse of the usual orientation).
Photos courtesy of Harry Noller, University of California, Santa Cruz.
 الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة


