النبات

مواضيع عامة في علم النبات

الجذور - السيقان - الأوراق

النباتات الوعائية واللاوعائية

البذور (مغطاة البذور - عاريات البذور)

الطحالب

النباتات الطبية


الحيوان

مواضيع عامة في علم الحيوان

علم التشريح

التنوع الإحيائي

البايلوجيا الخلوية


الأحياء المجهرية

البكتيريا

الفطريات

الطفيليات

الفايروسات


علم الأمراض

الاورام

الامراض الوراثية

الامراض المناعية

الامراض المدارية

اضطرابات الدورة الدموية

مواضيع عامة في علم الامراض

الحشرات


التقانة الإحيائية

مواضيع عامة في التقانة الإحيائية


التقنية الحيوية المكروبية

التقنية الحيوية والميكروبات

الفعاليات الحيوية

وراثة الاحياء المجهرية

تصنيف الاحياء المجهرية

الاحياء المجهرية في الطبيعة

أيض الاجهاد

التقنية الحيوية والبيئة

التقنية الحيوية والطب

التقنية الحيوية والزراعة

التقنية الحيوية والصناعة

التقنية الحيوية والطاقة

البحار والطحالب الصغيرة

عزل البروتين

هندسة الجينات


التقنية الحياتية النانوية

مفاهيم التقنية الحيوية النانوية

التراكيب النانوية والمجاهر المستخدمة في رؤيتها

تصنيع وتخليق المواد النانوية

تطبيقات التقنية النانوية والحيوية النانوية

الرقائق والمتحسسات الحيوية

المصفوفات المجهرية وحاسوب الدنا

اللقاحات

البيئة والتلوث


علم الأجنة

اعضاء التكاثر وتشكل الاعراس

الاخصاب

التشطر

العصيبة وتشكل الجسيدات

تشكل اللواحق الجنينية

تكون المعيدة وظهور الطبقات الجنينية

مقدمة لعلم الاجنة


الأحياء الجزيئي

مواضيع عامة في الاحياء الجزيئي


علم وظائف الأعضاء


الغدد

مواضيع عامة في الغدد

الغدد الصم و هرموناتها

الجسم تحت السريري

الغدة النخامية

الغدة الكظرية

الغدة التناسلية

الغدة الدرقية والجار الدرقية

الغدة البنكرياسية

الغدة الصنوبرية

مواضيع عامة في علم وظائف الاعضاء

الخلية الحيوانية

الجهاز العصبي

أعضاء الحس

الجهاز العضلي

السوائل الجسمية

الجهاز الدوري والليمف

الجهاز التنفسي

الجهاز الهضمي

الجهاز البولي


المضادات الميكروبية

مواضيع عامة في المضادات الميكروبية

مضادات البكتيريا

مضادات الفطريات

مضادات الطفيليات

مضادات الفايروسات

علم الخلية

الوراثة

الأحياء العامة

المناعة

التحليلات المرضية

الكيمياء الحيوية

مواضيع متنوعة أخرى

الانزيمات
Genes for rRNA Form Tandem Repeats Including an Invariant Transcription Unit
المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
المصدر:
LEWIN’S GENES XII
الجزء والصفحة:
18-3-2021
2668
Genes for rRNA Form Tandem Repeats Including an Invariant Transcription Unit
KEY CONCEPTS
-Ribosomal RNA (rRNA) is encoded by a large number of identical genes that are tandemly repeated to form one or more clusters.
-Each ribosomal DNA (rDNA) cluster is organized so that transcription units giving a joint precursor to the major rRNAs alternate with nontranscribed spacers.
-The genes in an rDNA cluster all have an identical sequence.
-The nontranscribed spacers consist of shorter repeating units whose number varies so that the lengths of individual spacers are different.
In the case of the globin genes discussed earlier, there are differences between the individual members of the cluster that allow selective pressure to act somewhat differently (but because of linkage, not independently) upon each gene. A contrast is provided by two cases of large gene clusters that contain many identical copies of the same gene or genes. Most eukaryotic organisms contain multiple copies of the genes for the histone proteins that are a major component of the chromosomes, and in most organismal genomes there are multiple copies of the genes that encode the ribosomal RNAs. These situations pose some interesting evolutionary questions.
Ribosomal RNA is the predominant product of transcription, constituting some 80% to 90% of the total mass of cellular RNA in both eukaryotes and prokaryotes. The number of major rRNA genes varies from 1 (in Coxiella burnetii, an obligate intracellular bacterium, and in Mycoplasma pneumoniae), to 7 in Escherichia coli, to 100 to 200 in unicellular/oligocellular eukaryotes, to several hundred in multicellular eukaryotes. The genes for the large and small rRNAs (found in the large and small subunits of the ribosome, respectively) usually form a tandem pair. (The sole exception is the yeast mitochondrion.)
The lack of any detectable variation in the sequences of the rRNA molecules implies that all of the copies of each gene must be identical. A point of major interest is what mechanism(s) are used to prevent variations from accumulating in the individual sequences.
In bacteria, the multiple rRNA genes are dispersed. In most eukaryotic genomes, the rRNA genes are contained in a tandem cluster or clusters. Sometimes these regions are called rDNA. (In some cases, the proportion of rDNA in the total DNA, together with its atypical base composition, is great enough to allow its isolation as a separate fraction directly from sheared genomic DNA.) An important diagnostic feature of a tandem cluster is that it generates a circular restriction map (see the Methods in Molecular Biology and Genetic Engineering chapter for a description of restriction mapping), as shown in FIGURE 1.
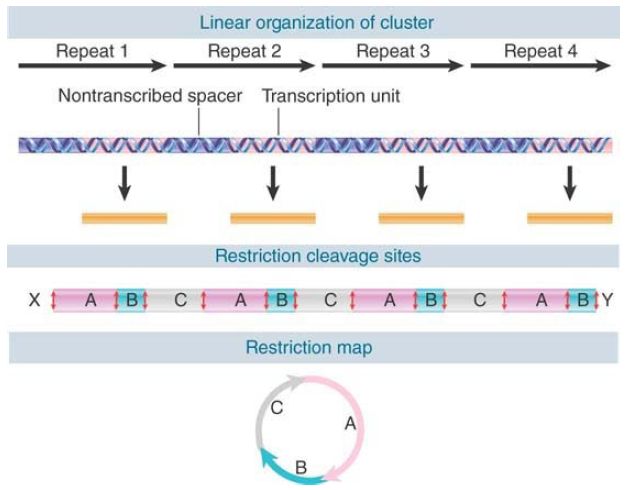
FIGURE 1 A tandem gene cluster has an alternation of transcription unit and nontranscribed spacer and generates a circular restriction map.
Suppose that each repeat unit has three restriction sites. When we map these fragments by conventional means, we find that A is next to B, which is next to C, which is next to A, generating the circular map. If the cluster is large, the internal fragments (A, B, and C) will be present in much greater quantities than the terminal fragments (X and Y) that connect the cluster to adjacent DNA. In a cluster of 100 repeats, X and Y would be present at 1% of the level of A, B, and C. This can make it difficult to obtain the ends of a gene cluster for mapping purposes.
The region of the nucleus where 18S and 28S rRNA synthesis occurs has a characteristic appearance, with a fibrillar core surrounded by a granular cortex. The fibrillar core is where the
rRNA is transcribed from the DNA template, and the granular cortex is formed by the ribonucleoprotein particles into which the rRNA is assembled. The entire area is called the nucleolus. Its characteristic morphology is evident in FIGURE 2.
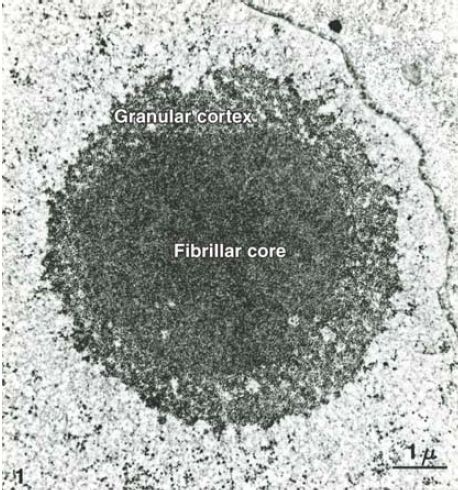
FIGURE 2. The nucleolar core identifies rDNA undergoing transcription and the surrounding granular cortex consists of assembling ribosomal subunits. This thin section shows the nucleolus of the newt Notophthalmus viridescens.
Photo courtesy of Oscar Miller.
The particular chromosomal regions associated with a nucleolus are called nucleolar organizers. Each nucleolar organizer corresponds to a cluster of tandemly repeated 18/28S rRNA genes on one chromosome. The concentration of the tandemly repeated rRNA genes, together with their very intensive transcription, is responsible for creating the characteristic morphology of the nucleoli.
The pair of major rRNAs is transcribed as a single precursor in both bacteria (where 5S and 16/23S rRNAs are cotranscribed) and the eukaryotic nucleolus (where the 18S and 28S rRNAs are transcribed). In eukaryotes, 5S genes are also typically found in tandem clusters transcribed as a precursor with transcribed spacers. Following transcription, the precursor is cleaved to release the individual rRNA molecules. The transcription unit is shortest in bacteria and is longest in mammals (where it is known as 45S RNA, according to its rate of sedimentation). An rDNA cluster contains many transcription units, each separated from the next by a nontranscribed spacer, so that many RNA polymerases are simultaneously engaged in transcription on one repeating unit.
The polymerases are so closely packed that the RNA transcripts form a characteristic matrix displaying increasing length along the transcription unit.
The length of the nontranscribed spacer varies a great deal between and (sometimes) within species. In yeast there is a short nontranscribed spacer that is relatively constant in length. In the fruit fly Drosophila melanogaster there is nearly twofold variation in the length of the nontranscribed spacer between different copies of the repeating unit. A similar situation is seen in the amphibian Xenopus laevis. In each of these cases, all of the repeating units are present as a single tandem cluster on one particular chromosome. (In the example of D. melanogaster, this happens to be the sex chromosomes. The cluster on the X chromosome is larger than the one on the Y chromosome, so female flies have more copies of the rRNA genes than male flies do.)
In mammals the repeating unit is much larger, comprising the transcription unit of about 13 kb and a nontranscribed spacer of about 30 kb. Usually, the genes lie in several dispersed clusters; in the cases of humans and mice the clusters reside on five and six chromosomes, respectively. One interesting question is how the corrective mechanisms that presumably function within a single cluster to ensure that rRNA copies are identical are able to work when there are several clusters. Recent research suggests that selection might maintain a coordinated number of functional copies of genes among clusters on different chromosomes to ensure that dosages of different rRNA molecules (which must interact in forming a ribosome) remain approximately equal.
The variation in length of the nontranscribed spacer in a single gene cluster contrasts with the conservation of sequence of the transcription unit. In spite of this variation, the sequences of longer nontranscribed spacers remain homologous with those of the shorter nontranscribed spacers. This implies that each nontranscribed spacer is internally repetitious, so that the variation in length results from changes in the number of repeats of some subunit.
The general nature of the nontranscribed spacer is illustrated by the example of X. laevis (FIGURE 3). Regions that are fixed in length alternate with regions that vary in length. Each of the three repetitive regions comprises a variable number of repeats of a rather short sequence. One type of repetitious region has repeats of a 97-bp sequence; the other, which occurs in two locations, has a repeating unit found in two forms, both 60 bp and 81 bp long. The variation in the number of repeating units in the repetitive regions accounts for the overall variation in spacer length. The repetitive regions are separated by shorter constant sequences called Bam islands. (This description takes its name from their isolation via the use of the BamHI restriction enzyme.) From this type of organization, we see that the cluster has evolved by duplications involving the promoter region.
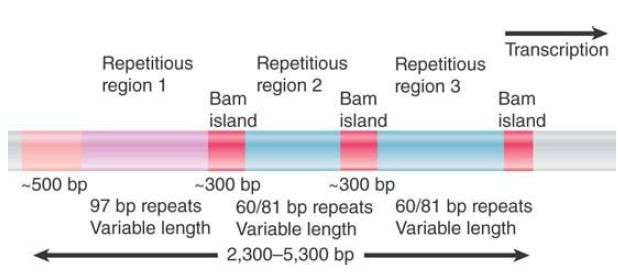
FIGURE 3 The nontranscribed spacer of X. laevis rDNA has an internally repetitious structure that is responsible for its variation in length. The Bam islands are short, constant sequences that
separate the repetitious regions.
We need to explain the lack of variation in the expressed copies of the repeated genes. One hypothesis would be that there is a quantitative demand for a certain number of “good” sequences.
However, this would enable mutated sequences to accumulate up to a point at which their proportion of the cluster is great enough for selection to act against them. We can exclude this hypothesis because of the lack of such variation in the cluster.
The lack of variation implies that there is negative selection against individual variations. Another hypothesis would be that the entire cluster is regenerated periodically from one or a very few members. As a practical matter, any mechanism would need to involve regeneration every generation. We can exclude this hypothesis because a regenerated cluster would not show variation in the nontranscribed regions of the individual repeats.
We are left with a dilemma. Variation in the nontranscribed regions suggests that there is frequent unequal crossing-over. This will change the size of the cluster but will not otherwise change the properties of the individual repeats. So, how are mutations prevented from accumulating? The following section shows that continuous contraction and expansion of a cluster might provide a mechanism for homogenizing its copies.
 الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة

الآخبار الصحية















 قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة
قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة "المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة
"المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة (نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)
(نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)