


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 18-11-2020
Date: 9-6-2021
Date: 30-12-2015
|
A Constant Rate of Sequence Divergence Is a Molecular Clock
KEY CONCEPTS
-The sequences of orthologous genes in different species vary at nonsynonymous sites (where mutations have caused amino acid substitutions) and synonymous sites (where mutation has not affected the amino acid sequence).
-Synonymous substitutions accumulate about 10 times faster than nonsynonymous substitutions.
-The evolutionary divergence between two DNA sequences is measured by the corrected percentage of positions at which the corresponding nucleotides differ.
-Substitutions can accumulate at a more or less constant rate after genes separate, so that the divergence between any pair of globin sequences is proportional to the time since they shared common ancestry.
Most changes in gene sequences occur by mutations that accumulate slowly over time. Point mutations and small insertions and deletions occur by chance, probably with more or less equal
probability in all regions of the genome. The exceptions to this are hotspots, where mutations occur much more frequently. Recall from the section DNA Sequences Evolve by Mutation and a Sorting Mechanism earlier in this chapter that most nonsynonymous mutations are deleterious and will be eliminated by negative selection, whereas the rare advantageous substitution will spread through the population and eventually replace the original sequence (fixation). Neutral variants are expected to be lost or fixed in the population due to random genetic drift. What proportion of mutational changes in a protein-coding gene sequence is selectively neutral is a historically contentious issue.
The rate at which substitutions accumulate is a characteristic of each gene, presumably depending at least in part on its functional flexibility with regard to change. Within a species, a gene evolves by mutation followed by fixation within the single population. Recall that when we study the genetic variation of a species, we see only the variants that have been maintained, whether by selection or genetic drift. When multiple variants are present they might be stable, or they might in fact be transient because they are in the process of being fixed (or lost).
When a single species separates into two new species, each of the resulting species constitutes an independent evolutionary lineage.
By comparing orthologous genes in two species, we see the differences that have accumulated between them since the time when their ancestors ceased to interbreed. Some genes are highly
conserved, showing little or no change from species to species. This indicates that most changes are deleterious and therefore eliminated.
The difference between two genes is expressed as their divergence, the percentage of positions at which the nucleotides are different, corrected for the possibility of convergent mutations (the same mutation at the same site in two separate lineages) and true revertants. There is usually a difference in the rate of evolution among the three codon positions within genes, because mutations at the third base position often are synonymous, as are some at the first position.
In addition to the coding sequence, a gene contains untranslated regions. Here again, most mutations are potentially neutral, apart from their effects on either secondary structure or (usually rather short) regulatory signals.
Although synonymous mutations are expected to be neutral with regard to the polypeptide, they could affect gene expression via the sequence change in RNA .
Another possibility is that a change in synonymous codons calls for a different tRNA to respond, influencing the efficiency of translation. Species generally show a codon bias; when there are multiple codons for the amino acid, one codon is found in protein-coding genes in a high percentage, whereas the remaining codons are found in low percentages. There is a corresponding percentage difference in the tRNA types that recognize these codons.
Consequently, a change from a common to a rare synonymous codon can reduce the rate of translation due to a lower concentration of appropriate tRNAs.
Researchers can measure the divergence of proteins (representing nonsynonymous changes in their genes) over time by comparing species for which there is paleontological evidence for the time of their divergence. Such data provide two general observations.
First, different proteins evolve at different rates. For example, fibrinopeptides evolve quickly, cytochrome c evolves slowly, and hemoglobin evolves at an intermediate rate. Second, for some proteins (including the three just mentioned), the rate of evolution is approximately constant over millions of years. In other words, for a given type of protein, the divergence between any pair of sequences is (more or less) proportional to the time since they shared a common ancestor. This provides a molecular clock that measures the accumulation of substitutions at an approximately constant rate during the evolution of a particular protein-coding gene.
There can also be molecular clocks for paralogous proteins diverging within a species lineage. To take the example of the human β- and δ-globin chains , there are 10 differences in 146 amino acids, a divergence of 6.9%. The DNA sequence has 31 changes in 441 nucleotides (7%). However, the nonsynonymous and synonymous changes are distributed very differently. There are 11 changes in the 330 nonsynonymous sites (3.3%), but 20 changes in only 111 synonymous sites (18%). This gives corrected rates of divergence of 3.7% in the nonsynonymous sites and 32% in the synonymous sites, an order of magnitude in difference.
The striking difference in the divergence of nonsynonymous and synonymous sites demonstrates the existence of much greater constraints on nucleotide changes that alter polypeptide sequences compared to those that do not. Many fewer amino acid changes are neutral.
Suppose that we take the rate of synonymous substitutions to indicate the underlying rate of mutational fixation (assuming there is no selection at all at the synonymous sites). Then, over the period since the β and δ genes diverged, there should have been changes at 32% of the 330 nonsynonymous sites, for a total of 105. All but 11 of them have been eliminated, which means that about 90% of the mutations were not retained.
The rate of divergence can be measured as the percent difference per million years or as its reciprocal, the unit evolutionary period (UEP)—the time in millions of years that it takes for 1% divergence to accumulate. After the rate of the molecular clock has been established by pairwise comparisons between species (remembering the practical difficulties in establishing the actual time since the existence of the common ancestor), it can be applied to paralogous genes within a species. From their divergence, we can calculate how much time has passed since the duplication that generated them.
By comparing the sequences of orthologous genes in different species, the rate of divergence at both nonsynonymous and synonymous sites can be determined, as plotted in FIGURE 1.
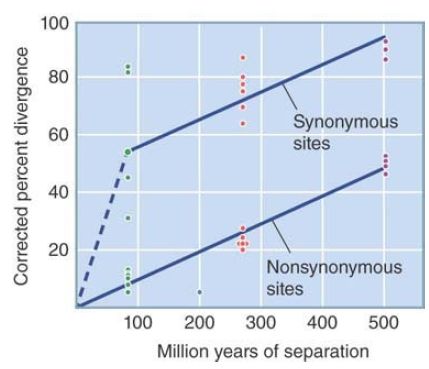
FIGURE 1 Divergence of DNA sequences depends on evolutionary separation. Each point on the graph represents a pairwise comparison.
In pairwise comparisons, there is an average divergence of 10% in the nonsynonymous sites of either the α- or β-globin genes of mammal lineages that have been separated since the mammalian radiation occurred roughly 85 million years ago. This corresponds to a nonsynonymous divergence rate of 0.12% per million years.
The rate is approximately constant when the comparison is extended to genes that diverged in the more distant past. For example, the average nonsynonymous divergence between orthologous mammalian and chicken globin genes is 23%. Relative to a common ancestor at roughly 270 million years ago, this gives a rate of 0.09% per million years.
Going farther back, we can compare the α- with the β-globin genes within a species. They have been diverging since the original duplication event about 500 million years ago (see FIGURE 2). They have an average nonsynonymous divergence of about 50%, which gives a rate of 0.1% per million years.
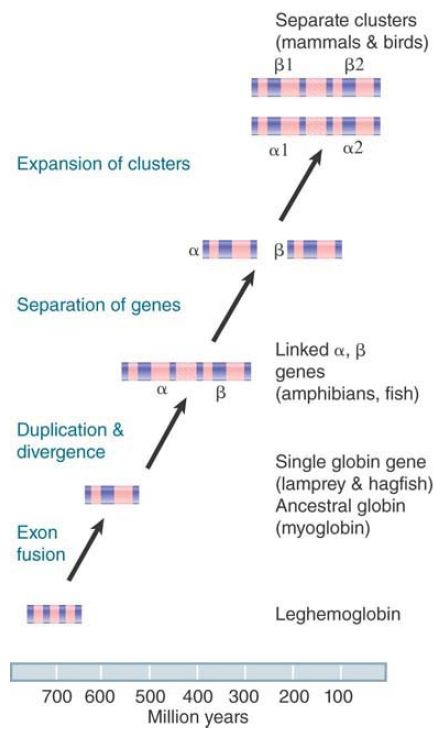
FIGURE 2. All globin genes have evolved by a series of duplications, transpositions, and mutations from a single ancestral gene.
The summary of these data in Figure 1 shows that nonsynonymous divergence in the globin genes has an average rate of about 0.096% per million years (for a UEP of 10.4). Considering the uncertainties in estimating the times at which the species diverged, the results lend good support to the idea that there is a constant molecular clock.
The data on synonymous site divergence are much less clear. In every case, it is evident that the synonymous site divergence is much greater than the nonsynonymous site divergence, by a factor that varies from 2 to 10. However, the range of synonymous site divergences in pairwise comparisons is too great to establish a molecular clock, so we must base temporal comparisons on the nonsynonymous sites.
From Figure 1, it is clear that the rate of evolution at synonymous sites is only approximately constant over time. If we assume that there must be zero divergence at zero years of separation, we see that the rate of synonymous site divergence is much greater for the first approximately 100 million years of separation. One interpretation is that roughly half of the synonymous sites are rapidly (within 100 million years) saturated by mutations; this half behaves as neutral sites. The other half accumulates mutations more slowly, at a rate approximately the same as that of the nonsynonymous sites; this half represents sites that are synonymous with regard to the polypeptide but that are under selective constraint for some other reason.
Now we can reverse the calculation of divergence rates to estimate the times since paralogous genes were duplicated. The difference between the human β and α genes is 3.7% for nonsynonymous sites. At a UEP of 10.4, these genes must have diverged 10.4 ×3.7 = about 40 million years ago—about the time of the separation of the major primate lineages: New World monkeys, Old World monkeys, and great apes (including humans). All of these taxonomic groups have both β and δ genes, which suggests that the gene divergence began just before this point in evolution.
Proceeding further back, the divergence between the nonsynonymous sites of γ and ε genes is 10%, which corresponds to a duplication event about 100 million years ago. The separation between embryonic and fetal globin genes therefore might have just preceded or accompanied the mammalian radiation.
An evolutionary tree for the human globin genes is presented in FIGURE 3. Paralogous groups that evolved before the mammalian radiation—such as the separation of β/δ from γ—
should be found in all mammals. Paralogous groups that evolved afterward—such as the separation of β- and δ-globin genes—should be found in individual lineages of mammals.
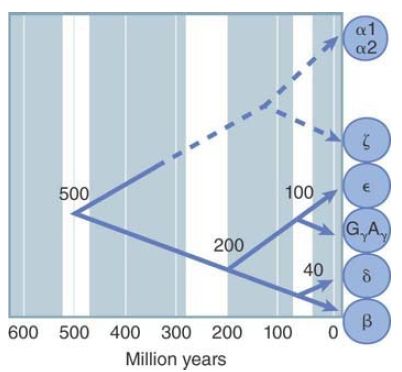
FIGURE 3 Nonsynonymous site divergences between pairs of ββ-globin genes allow the history of the human cluster to be reconstructed. This tree accounts for the separation of classes of globin genes.
In each species, there have been comparatively recent changes in the structures of the clusters. We know this because we see differences in gene number (one adult β-globin gene in humans, two in the mouse) or in type (most often concerning whether there are separate embryonic and fetal genes).
When sufficient data have been collected on the sequences of a particular gene or gene family, the analysis can be reversed and comparisons between orthologous genes can be used to assess
taxonomic relationships. If a molecular clock has been established, the time to common ancestry between the previously analyzed species and a species newly introduced to the analysis can be estimated.

|
|
|
|
دراسة: إجراء واحد لتقليل المخاطر الجينية للوفاة المبكرة
|
|
|
|
|
|
|
"الملح والماء" يمهدان الطريق لأجهزة كمبيوتر تحاكي الدماغ البشري
|
|
|
|
|
|
قسم شؤون المعارف يدعو الباحثين إلى المشاركة في مؤتمر علمي حول السيدة الزهراء (عليها السلام)
|
|
|
|
قسم العلاقات العامة يقدم ورشة ثقافية لمنسقي فتية الكفيل في جامعة القادسية
|
|
|
|
قسم الشؤون الفكرية يشارك في احتفالية تكريم الطلبة المتفوقين في العاصمة بغداد
|
|
|
|
جولات تعريف عن فعاليات المؤتمر العلمي الدولي السنوي الثالث عشر
|